Precision Medicine: The Solution to Finding Cures to Cancer?
By: Alyssa Chen
Breast cancer is one of the most common cancers in women worldwide, and is the second leading cause of death for women (Sun 2017). It is estimated that 1.7 new million cases are diagnosed every year worldwide (Stewart, 2014), approximately 570,000 of which lead to death (Sun, 2017). As a complex disease, breast cancer contains a wide variety of subtypes characterized by differences at molecular and cellular levels. A molecular subtype constituting 10-20% of all breast cancers (Lehman 2016), Triple Negative Breast Cancer (TNBC), lacks estrogen receptor and progesterone receptor (ER/PR) and human epidermal growth factor receptor 2 (HER2) expression (Bayraktar 2013). While other breast cancer subtypes have seen significant improvements in researched therapies, TNBC treatments have only recently started to gain ground. Compared to other breast cancers, TNBC tumors are generally larger and more biologically aggressive (Hafty 2006); furthermore, research finds 30% of metastatic TNBC patients survive to 5 years, with almost all dying despite receiving adjuvant chemotherapy (Dent 2007). TNBC’s varying morphology, gene expression, and signaling pathway activity contribute to its characterization as high-heterogeneity disease (Wu 2018). Thus, the inability to have a “one size fits all” approach to treating TNBC complicates prognosis and treatment.
Because endocrine agents have proven ineffective, cytotoxic chemotherapy currently serves as the standard treatment for TNBC patients (Hossain 2021). However, a new field is emerging: precision medicine. Rather than treating all TNBC patients using universal treatment practices, precision medicine utilizes patient genomic information to identify the most accurate and efficient treatment on a patient-by-patient basis. This way, treatment is tailored to the individual and not applied to a field of patients with varying genetic characteristics who may each respond differently to a universally-applied treatment. In this article, we discuss ways precision medicine can treat TNBC, specifically with regard to molecular subtyping and investigation of oncogenic signaling pathway irregularities attributed to TNBC.
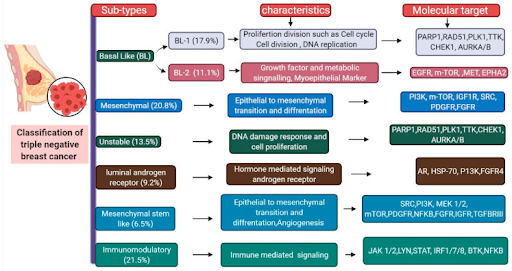
Molecular subtyping enables researchers to effectively isolate and study TNBC subtypes with shared genetic characteristics, and consequently devise subtype-based treatments with greater accuracy. While TNBC itself is a molecular subtype of breast cancer, its many varying characteristics can be categorized utilizing further molecular subtyping practices. Researchers can compile gene expression (GE) profiles from publicly available datasets or their own clinical trials, and through statistical technique and analysis, identify patterns and shared characteristics between data, yielding different molecular subtypes. Some subtypes by researchers include Burstein et al.’s classification of TNBC into 4 subtypes: luminal androgen receptor (LAR), mesenchymal (M), basal-like immunosuppressed (BLIS), and basal-like immune-activated (BLIA) (Burstein, 2015). Although since reclassified, in 2011, Lehmann et al. arranged TNBC into 6 subtypes : basal-like 1 (BL-1), basal-like 2 (BL-2), M, Mesenchymal stem-like (MSL), Immunomodulatory (IM), and LAR (Hossain, 2021). This study defied previous misconceptions regarding TNBC’s tumors as exhibiting only basal-like phenotypes. Furthermore, subtype classification enabled isolated drug research and development for each subtype. For BL1 and BL2 subtypes, which exhibit high gene expression levels in cell proliferation and DNA damage response, Lehmann et al. proposed and successfully showed anti-mitotic and DNA-damaging agents as promising treatment for this subtype. (Lehmann 2011). Although different TNBC subtype groups and names are researcher-dependent, the universal effect of subtyping is that it more efficiently organizes TNBC into categories to help isolate and identify treatment efficiency.
Figure 1. TNBC molecular subtypes with respective characteristics and molecular targets. Image from Singh 2021.
In addition to subtyping, researchers also look to oncogenic signaling pathways associated with TNBC to identify potential targeted treatments. For example, dysregulation in the P13K/AKT/mTOR pathway commonly occurs in TNBC, with P13KCA-gain of function mutations present in 23.7% of TNBC patients (Cossu-Rocca 2017). Researchers use their knowledge of the P13K/AKT/mTOR pathway to develop pharmaceutical drugs able to target or minimize the dysregulated pathway. Preclinical results presented inhibition of mammalian target of rapamycin (mTOR) as promising treatment, leading to the creation and FDA-approval of Everolimus, a selective mTOR inhibitor with the ability to inhibit tumor cell growth and proliferation and indirectly inhibit angiogenesis (Singh 2014). Other molecular agents such as cyclin-dependent kinase (CDK) inhibitors have shown promising results but still remain in preclinical phase trials. To curb abnormal CDK expression commonly present in TNBC, inhibitors (primarily CDK 4/6) amplify anti-tumor immunity through T-cell activation; thus, many CDK inhibitors are tested in combination with immunotherapy drugs(Deng 2018). Inhibitors currently in phase 1 clinical trials include Dinaciclib, a pan-CDK inhibitor undergoing trial in combination with pembrolizumab, and Trilaciclub, a CDK 4/6 Inhibitor undergoing trail in combination with gemcitabine and carboplatin (Hossain 2021). While these and other developing drug combinations have yet to gain approval, successful preclinical results show their promising futures for FDA-approved use in clinical settings.
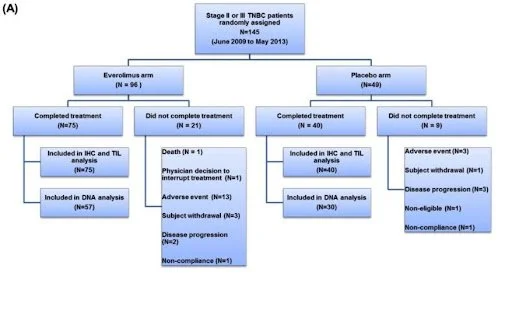
Figure 2. Example of Consort Diagram of Patient Flow from Jovanović et al. Everolimus-efficacy study. Image from Jovanović 2017.
Although exhibiting promising benefits, precision medicine does not present without drawbacks. As a more novel approach to treating TNBC, further research is required to better understand precision oncology and its long-term effects. In regards to molecular subtyping practices, further efforts are needed to improve research-study-design, as TNBC’s high variability in pathogenic genes causing TNBC often results in small sample size per molecular subtype; therefore, the promising drugs emerging from studies are only applicable to a small number of patients, and further drug research and development is necessary for many patients with other subtypes. (Zhang 2019). Moreover, the evolution of selective pressure and new mutations within tumors makes potential drug treatments effective only at a certain space and time (Zhang 2019). Additionally, as it currently stands, inaccessibility to precision medicine, particularly in low-income countries, remains an issue for ensuring equal accessibility for all patients (Hamdi 2011). Various nations have demonstrated efforts to expand access, including Obama’s NIH-funded Precision Medicine Initiative in 2015 (Hossain 2021) and Sweden’s successful Sweden Cancerome Analysis Network - Breast (SWAT-B) Initiative (Ryden 2018), setting precedents for potential application in other nations or at international levels.
As the world searches for “the cure for cancer”, precision medicine is emerging as a promising solution. Through molecular subtyping and oncogenic signal pathway targeting, its recent successes in discovering different subtype- or pathway-based targeted therapies have led to discovery of treating patients with an individualized, and thus, more precise, approach. As research within it expands and evolves, precision medicine could be the answer to finding the cures of cancer - resulting in more accurate and effective treatment for all.
References
Burstein, M. D., Tsimelzon, A., Poage, G. M., Covington, K. R., Contreras, A., Fuqua, S. A., Savage, M. I., Osborne, C. K., Hilsenbeck, S. G., Chang, J. C., Mills, G. B., Lau, C. C., & Brown, P. H. (2015). Comprehensive genomic analysis identifies novel subtypes and targets of triple-negative breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research, 21(7), 1688–1698. https://doi.org/10.1158/1078-0432.CCR-14-0432
Cossu-Rocca, P., Orrù, S., Muroni, M. R., Sanges, F., Sotgiu, G., Ena, S., Pira, G., Murgia, L., Manca, A., Uras, M. G., Sarobba, M. G., Urru, S., & De Miglio, M. R. (2015). Analysis of PIK3CA Mutations and Activation Pathways in Triple Negative Breast Cancer. PloS one, 10(11), e0141763. https://doi.org/10.1371/journal.pone.0141763
Deng, J., Wang, E. S., Jenkins, R. W., Li, S., Dries, R., Yates, K., Chhabra, S., Huang, W., Liu, H., Aref, A. R., Ivanova, E., Paweletz, C. P., Bowden, M., Zhou, C. W., Herter-Sprie, G. S., Sorrentino, J. A., Bisi, J. E., Lizotte, P. H., Merlino, A. A., Quinn, M. M., … Wong, K. K. (2018). CDK4/6 Inhibition Augments Antitumor Immunity by Enhancing T-cell Activation. Cancer discovery, 8(2), 216–233. https://doi.org/10.1158/2159-8290.CD-17-0915
Dent, R., Trudeau, M., Pritchard, K. I., Hanna, W. M., Kahn, H. K., Sawka, C. A., Lickley, L. A., Rawlinson, E., Sun, P., & Narod, S. A. (2007). Triple-negative breast cancer: clinical features and patterns of recurrence. Clinical cancer research : an official journal of the American Association for Cancer Research, 13(15 Pt 1), 4429–4434. https://doi.org/10.1158/1078-0432.CCR-06-3045
Haffty, B. G., Yang, Q., Reiss, M., Kearney, T., Higgins, S. A., Weidhaas, J., Harris, L., Hait, W., & Toppmeyer, D. (2006). Locoregional relapse and distant metastasis in conservatively managed triple negative early-stage breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology, 24(36), 5652–5657. https://doi.org/10.1200/JCO.2006.06.5664
Hamdi, Y., Mighri, N., Boujemaa, M., Mejri, N., Ben Nasr, S., Ben Rekaya, M., Messaoud, O., Bouaziz, H., Berrazega, Y., Rachdi, H., Jaidane, O., Daoud, N., Zribi, A., Ayari, J., El Benna, H., Labidi, S., Ben Hassouna, J., Haddaoui, A., Rahal, K., Benna, F., … Abdelhak, S. (2021). Identification of Eleven Novel BRCA Mutations in Tunisia: Impact on the Clinical Management of BRCA Related Cancers. Frontiers in oncology, 11, 674965. https://doi.org/10.3389/fonc.2021.674965
Hossain, F., Majumder, S., David, J., & Miele, L. (2021). Precision Medicine and Triple-Negative Breast Cancer: Current Landscape and Future Directions. Cancers, 13(15), 3739. https://doi.org/10.3390/cancers13153739
Lehmann, B. D., Bauer, J. A., Chen, X., Sanders, M. E., Chakravarthy, A. B., Shyr, Y., & Pietenpol, J. A. (2011). Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. The Journal of clinical investigation, 121(7), 2750–2767. https://doi.org/10.1172/JCI45014
Rydén, L., Loman, N., Larsson, C., Hegardt, C., Vallon-Christersson, J., Malmberg, M., Lindman, H., Ehinger, A., Saal, L. H., & Borg, Å. (2018). Minimizing inequality in access to precision medicine in breast cancer by real-time population-based molecular analysis in the SCAN-B initiative. The British journal of surgery, 105(2), e158–e168. https://doi.org/10.1002/bjs.10741
McGuire S. (2016). World Cancer Report 2014. Geneva, Switzerland: World Health Organization, International Agency for Research on Cancer, WHO Press, 2015. Advances in nutrition (Bethesda, Md.), 7(2), 418–419. https://doi.org/10.3945/an.116.012211
Singh, J., Novik, Y., Stein, S., Volm, M., Meyers, M., Smith, J., Omene, C., Speyer, J., Schneider, R., Jhaveri, K., Formenti, S., Kyriakou, V., Joseph, B., Goldberg, J. D., Li, X., Adams, S., & Tiersten, A. (2014). Phase 2 trial of everolimus and carboplatin combination in patients with triple negative metastatic breast cancer. Breast cancer research : BCR, 16(2), R32. https://doi.org/10.1186/bcr3634
Sun, Y. S., Zhao, Z., Yang, Z. N., Xu, F., Lu, H. J., Zhu, Z. Y., Shi, W., Jiang, J., Yao, P. P., & Zhu, H. P. (2017). Risk Factors and Preventions of Breast Cancer. International journal of biological sciences, 13(11), 1387–1397. https://doi.org/10.7150/ijbs.21635
Wu, N., Zhang, J., Zhao, J., Mu, K., Zhang, J., Jin, Z., Yu, J., & Liu, J. (2018). Precision medicine based on tumorigenic signaling pathways for triple-negative breast cancer. Oncology letters, 16(4), 4984–4996. https://doi.org/10.3892/ol.2018.9290
Zhang, X., Yang, H., & Zhang, R. (2019). Challenges and future of precision medicine strategies for breast cancer based on a database on drug reactions. Bioscience reports, 39(9), BSR20190230. https://doi.org/10.1042/BSR20190230
Images
Jovanović, B., Mayer, I. A., Mayer, E. L., Abramson, V. G., Bardia, A., Sanders, M. E., Kuba, M. G., Estrada, M. V., Beeler, J. S., Shaver, T. M., Johnson, K. C., Sanchez, V., Rosenbluth, J. M., Dillon, P. M., Forero-Torres, A., Chang, J. C., Meszoely, I. M., Grau, A. M., Lehmann, B. D., … Pietenpol, J. A. (2017). A randomized phase II neoadjuvant study of cisplatin, paclitaxel with or without everolimus in patients with stage II/III triple-negative breast cancer (TNBC): Responses and long-term outcome correlated with increased frequency of DNA damage response gene mutations, TNBC subtype, AR status, and KI67. Clinical Cancer Research. Retrieved May 23, 2022. From https://pubmed.ncbi.nlm.nih.gov/28270498/
Singh, D. D., & Yadav, D. K. (2021). TNBC: Potential Targeting of Multiple Receptors for a Therapeutic Breakthrough, Nanomedicine, and Immunotherapy. Biomedicines, 9(8), 876. Retrieved April 8, 2022. From https://doi.org/10.3390/biomedicines9080876